Causal survival analysis
What If: Chapter 17
Elena Dudukina
2022-03-23
17.1 Hazards and risks
- Average causal effect of smoking cessation on the time to death
- The outcome is time to an event of interest that can occur at any time after the start of follow- up
- Administrative end of follow-up
- Competing events
- If the competing event is not a censoring event, then the analysis sets the time to event to be infinite
- If the competing event is a censoring event, then the analysis is an attempt to simulate a population in which death from other causes is abolished
- Create a composite event that includes both the competing event and the event of interest (e.g., death and stroke)
- Eliminates the competing events, but fundamentally changes the causal question
- Restrict the inference to the principal stratum of individuals who would not die regardless of the treatment level they receive
- Interpretation and valid estimation are hard
- None of the above strategies provides a satisfactory solution to the problem of competing events
17.1 Hazards and risks
- Survival probability
- Pr[T>k]
- Risk
- Cumulative incidence, at time k as one minus the survival: 1−Pr[T>k]=Pr[T<=K
- Hazard
- The proportion of individuals at any time k who develop the event among those who had not developed it before k
- Pr[T=k|T>k−1]
Data
library(readxl)temp <- tempfile()temp2 <- tempfile()download.file("https://cdn1.sph.harvard.edu/wp-content/uploads/sites/1268/2017/01/nhefs_excel.zip", temp)unzip(zipfile = temp, exdir = temp2)data <- read_xls(file.path(temp2, "NHEFS.xls"))unlink(c(temp, temp2))library(tidyverse)library(magrittr)# define/recode variables data %<>% mutate( # censoring cens = if_else(is.na(wt82_71), 1, 0), # categorize the school variable (as in R code by Joy Shi and Sean McGrath) education = cut(school, breaks = c(0, 8, 11, 12, 15, 20), include.lowest = TRUE, labels = c('1. 8th Grage or Less', '2. HS Dropout', '3. HS', '4. College Dropout', '5. College or More')), active = factor(active), exercise = factor(exercise), survtime = if_else(death == 0, 120, (yrdth-83)*12 + modth) # yrdth ranges from 83 to 92 ) %>% filter(!is.na(education))The hazards of hazard ratios
- The hazards vary over time; the hazard ratio generally does as well
- "Single hazard ratio, which is usually the consequence of fitting a Cox proportional hazards model that assumes a constant hazard ratio by ignoring interactions with time"
- Reported hazard ratio is a weighted average of the k-specific hazard ratios
- Presenting the time-specific hazard ratios also do not have straightforward causal interpretation
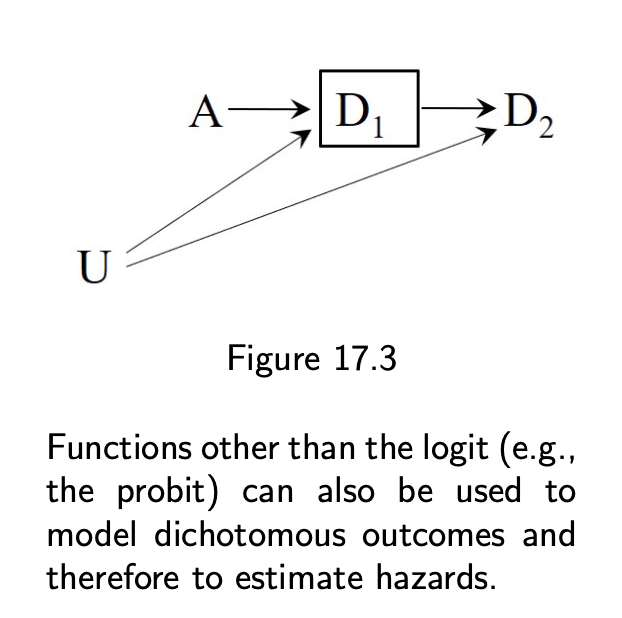
- Conditioning on the collider D1 will generally open the path A → D1 ← U → D2 and therefore induce an association between treatment A and event D2 among those with D1 = 0
Models for survival analysis
- Accommodate the expected censoring of failure times due to administrative end of follow-up
- Competing risk
- Constructing nonparametric Kaplan-Meier curves means no assumptions about the distribution of the unobserved failure times due to administrative censoring
- Parametric models for survival analysis assume a statistical distribution (e.g., exponential, Weibull) for the failure times or hazards
- Semiparametric Cox proportional hazards model and the accelerated failure time (AFT) model, do not assume a particular distribution for the failure times or hazards
Agnostic about the shape of the hazard when all covariates in the model have value of zero–often referred to as the baseline hazard
Assume restrictions on the relation between the baseline hazard and the hazard under other combinations of covariate values
Approximating the hazard ratio via a logistic model
Discrete time HR
Pr[Dk+1=1|Dk=0,A=1]Pr[Dk+1=1|Dk=0,A=0] is exp(α1) at all times k+1 in the hazards model Pr[Dk+1=1|Dk=0,A]=Pr[Dk+1|Dk=0,A=0]exp(α1A)
log(Pr[Dk+1=1|Dk=0,A])=α0,k+α1k, where Pr[Dk+1=1|Dk=0,A=0]=α0,k
When k+1 is small ➡️ Pr[Dk+1=1|Dk=0,A=0]≈0, the he hazard is ≈ to the odds
Odds: Pr[Dk+1=1|Dk=0,A]Pr[Dk+1=0|Dk=0,A]
log(Pr[Dk+1=1|Dk=0,A]Pr[Dk+1=0|Dk=0,A]) = logit(Pr[Dk+1=1|Dk=0,A]) = α0,k+α1k
If the hazard is close to zero at k+1, one can approximate the log hazard ratio by a coefficient from a logistic model
- Need to define a time unit k that is short enough for Pr[Dk+1=1|Dk=0,A]<0.1 to hold
17.3 Why censoring matters
- Not ready to assume that censoring is as if randomly assigned
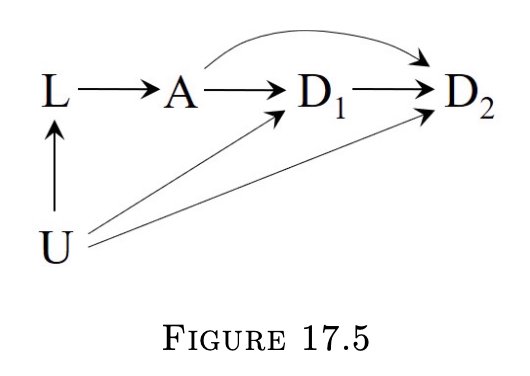
17.4 IP weighting of marginal structural models
- In the situation of no exchageability between the treated and untreated, direct contrast of their survival curves is not warranted
- L is sufficient for conditional exchangeability
- Assume positivity and consistency
17.4 IP weighting of marginal structural models
- Estimate the stabilized IP weight
ipw_num <- glm(qsmk ~ 1, data = data, family = binomial(link = "logit"))# denominator (propensity score model for the treatment)ipw_denom <- glm(qsmk ~ sex + race + age + I(age*age) + as.factor(education) + smokeintensity + I(smokeintensity*smokeintensity) + smokeyrs + I(smokeyrs*smokeyrs) + as.factor(exercise) + as.factor(active) + wt71 + I(wt71*wt71), data = data, family = binomial(link = "logit"))data %<>% mutate( # predicted probability of the treatment p_x = predict(ipw_num, type = "response"), p_x = case_when( qsmk == 1 ~ p_x, is.na(qsmk) ~ NA_real_, TRUE ~ 1 - p_x ), # predicted probability of the treatment given confounders p_x_l = predict(ipw_denom, type = "response"), p_x_l = case_when( qsmk == 1 ~ p_x_l, is.na(qsmk) ~ NA_real_, TRUE ~ 1-p_x_l ), # stabilized IPTW iptw_stab = p_x / p_x_l)17.4 IP weighting of marginal structural models
- Using the person-time data format, fit a hazards model
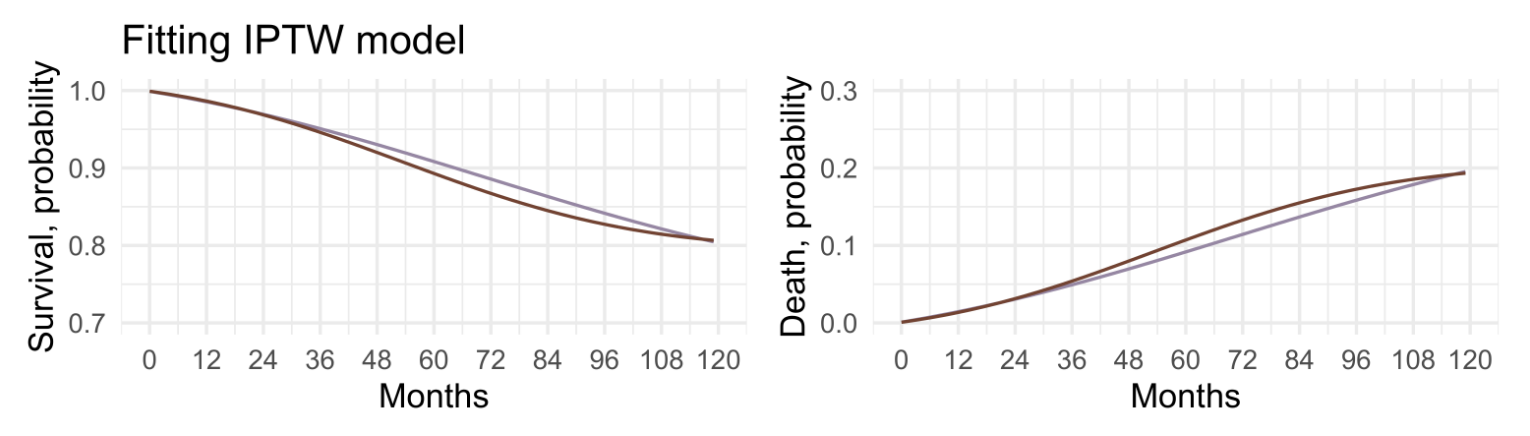
The IP weighted model estimates the time-varying hazards that would have been observed if all individuals in the study population had been treated (a = 1) and the time-varying hazards if they had been untreated (a = 0)
- Plotting code is available here
17.4 IP weighting of marginal structural models
Though the survival curve under treatment was lower than the curve under no treatment for most of the follow-up, the maximum difference never exceeded −1.4% with a 95% confidence interval from −3.4% to 0.7%.
Little evidence of an effect of smoking cessation on mortality at any time during the follow-up.
17.5 The parametric g-formula
- Step 1: Fit a parametric hazards model
# expand original data set until the last observed month for everyonemonths_gform <- data %>% # using observed data and observed censoring times uncount(weights = survtime, .remove = F) %>% group_by(seqn) %>% mutate ( time = row_number() - 1, event = case_when( time == survtime -1 & death == 1 ~ 1, TRUE ~ 0 ), timesq = time*time ) %>% ungroup() %>% select(seqn, qsmk, time, timesq, age, sex, race, education, smokeintensity, smkintensity82_71, smokeyrs, exercise, active, wt71, event)# fitting Q-model (model for the outcome) with confounders and outcome predictors for better precision in observation-month dataq_model <- glm(event == 0 ~ qsmk + I(qsmk*time) + I(qsmk*timesq) + time + timesq + sex + race + age + I(age*age) + as.factor(education) + smokeintensity + I(smokeintensity*smokeintensity) + smkintensity82_71 + smokeyrs + I(smokeyrs*smokeyrs) + as.factor(exercise) + as.factor(active) + wt71 + I(wt71*wt71), data = months_gform, family = binomial(link = "logit"))17.5 The parametric g-formula
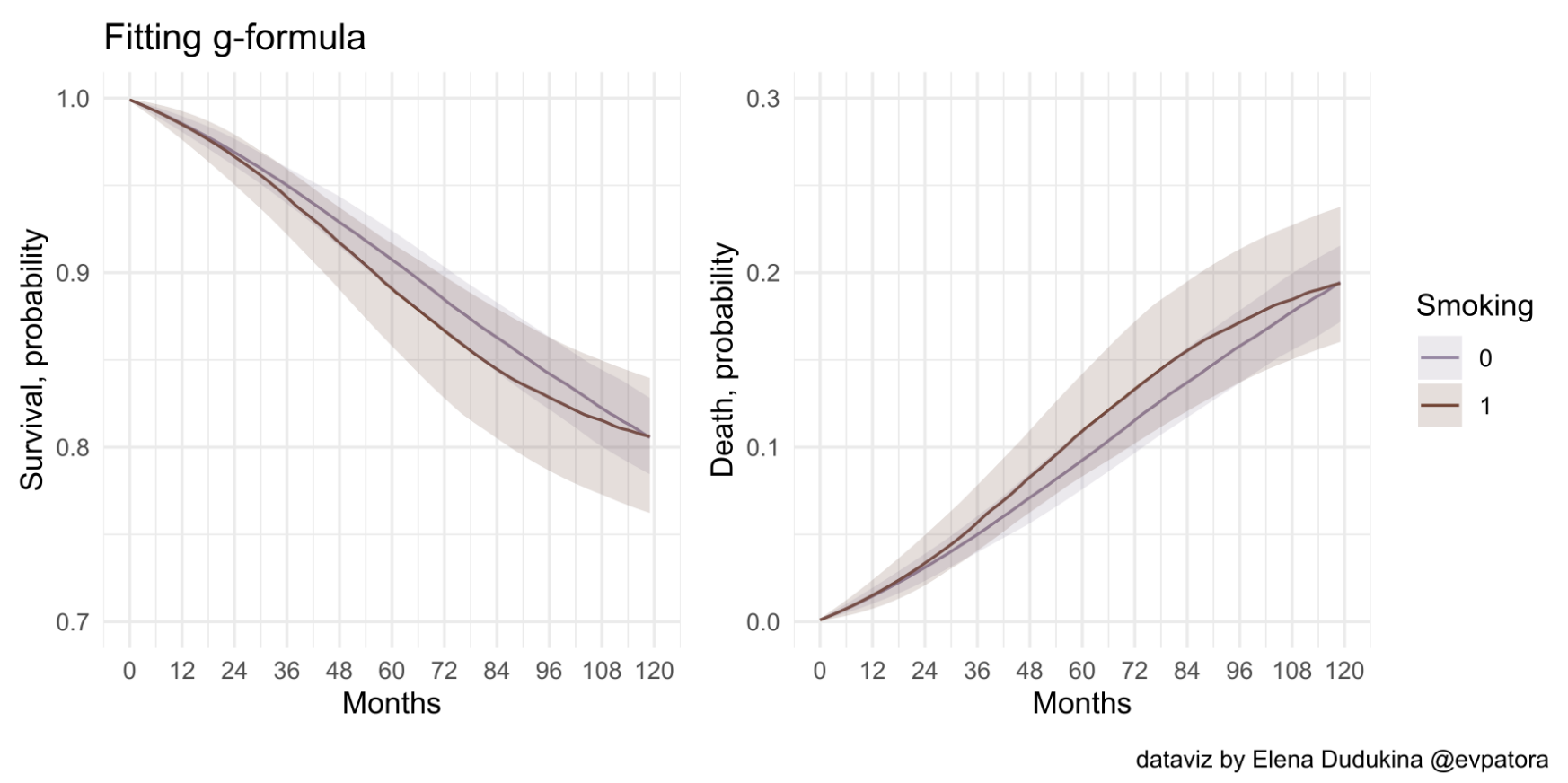
- Step 2: Compute the weighted average of the conditional survival across all values l of the covariates L (standardizing the survival to the confounder distribution)
- Plotting code is available here
17.5 The parametric g-formula
After adjustment for the covariates L via standardization, we found little evidence of an effect of smoking cessation on mortality at any time during the follow-up. Note that the survival curves estimated via IP weighting and the parametric g-formula are similar but not identical because they rely on different parametric assumptions.
The IP weighted estimates require no misspecification of a model for treatment and a model for the unconditional hazards.
The parametric g-formula estimates require no misspecification of a model for the conditional hazards.
17.6 G-estimation of structural nested models
Structural nested log-linear model to model the ratio of cumulative incidences (i.e., risks) under different treatment levels
- Structural nested cumulative failure time model
- Used when survival is rare because log-linear models do not naturally impose an upper limit of 1 on the survival
Accelerated failure time (AFT) model
- Models the ratio of survival times under different treatment options
- Rarely been used in practice due to the lack of user-friendly software
- Parameters of structural AFT models estimated via search algorithms do not guarantee to find a unique solution
References
- Hernán MA, Robins JM (2020). Causal Inference: What If. Boca Raton: Chapman & Hall/CRC (v. 30mar21)
- Causal Survival Analysis: IPTW and 95% CI bands
- Causal Survival Analysis: Parametric g-formula and and 95% CI bands